Langen Lab: Amyloid and Membrane Protein Structure and Function

| Email: | langen@usc.edu |
| Office Location: | ZNI room 119 |
| Office Phone: | (323) 442 1323 |
| Lab Location: | ZNI 121 |
| Lab Phone: | (323) 442 0106 |
| Ph.D. Programs: | PIBBS |
Our main interests lie in the basic areas of protein folding and misfolding, and particular emphasis is placed on these processes when they occur on or around membranes. The interaction of proteins with membranes underlies many important biological processes. Proteins can regulate the structure and function of biological membranes by controlling the composition, fluidity, permeability and curvature of cellular membranes. Membranes, in turn, can have a pronounced effect on the structure and function of proteins and help to promote physiologically important structural reorganizations of proteins. In addition, membrane interaction can also result in protein misfolding which could ultimately cause disease. We investigates the interaction between proteins and membranes in two biological contexts:
- Recognition and stabilization of membrane curvature. Although regulation of membrane curvature is a key event in many biological processes, including endocytosis, exocytosis and vesicle budding, it remains poorly understood. We are studying structural features that allow proteins to sense or stabilize membrane curvature in endocytosis.
- Protein misfolding and amyloid fibril formation in disease. We have taken an interest in the areas of protein misfolding and amyloid fibril formation. We are particularly interested in protein misfolding as it pertains to Alzheimer's disease, Parkinson's disease, Huntington's disease and type-2 diabetes. It is our objective to understand how certain misfolded forms of amyloid proteins are able to insert into membranes and perturb membrane structure. Furthermore, we are investigating both the general molecular mechanisms that cause amyloid fibril formation and the manner by which this process is modulated by phospholipids.
Despite significant interest and biomedical relevance, it has proven difficult to address these questions using conventional high-resolution structural methods. We have therefore been applying a broad range of biophysical methods. To date, site-directed spin labeling (SDSL), in combination with electron paramagnetic resonance (EPR) spectroscopy, has been most instrumental in our endeavors. SDSL is a powerful new approach for the structural analysis of membrane proteins, and, as we recently found, amyloid fibrils. The basis of SDSL is to introduce a spin label into a given protein sequence. Through EPR analysis of multiple derivatized proteins, it is possible to determine local structure and dynamics, to map inter-residue distances, and, ultimately, to generate reliable three-dimensional structures.
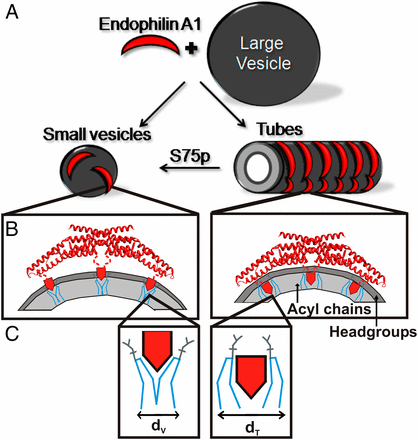
Schematic illustration of endophilin A1 tube and vesicle binding and its modulation by phosphorylation.
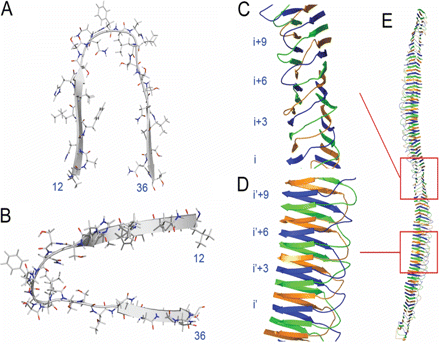
Model of hIAPP fibril structure

