Rohs Lab: Computational Structural Biology

| Email: | rohs AT usc.edu |
| Office Location: | RRI 413H |
| Office Phone: | (213) 740 0552 |
| Website | http://rohslab.cmb.usc.edu/ |
| Ph.D. Programs: | Computational Biology and Bioinformatics, Molecular Biology, Chemistry, Physics, and Computer Science. |
The main focus of our Computational Structural Biology research group is the integration of two fields of research: genomics and structural biology. Our primary goal is to reveal yet unknown molecular mechanisms underlying gene regulation using bioinformatics analyses of high-throughput sequencing and DNA methylation data of whole genomes.
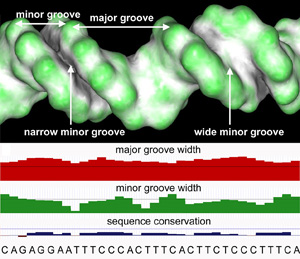
Genome analysis views DNA as a linear string of the letters A, C, G, and T but proteins recognize DNA as a three-dimensional object (see Figure). Our main interest is to understand better how transcription factors (TFs) recognize nuances in intrinsic DNA structure and to identify TF families for which the readout of local DNA shape contributes to binding specificity and explains distinct functions of closely related TFs. Until recently, our research mainly focused on the analysis of TF binding sites (TFBSs) for which structural information was available. However, sequence information for whole genomes has become available in recent years due to advances in high-throughput sequencing technologies, whereas structural information on that scale is not available. It is still unknown why certain TFs bind to similar DNA sequences but execute different in vivo functions, or in turn bind to diverse sequences. Our scientific contributions suggest that direct chemical contacts with base pairs cannot sufficiently explain binding specificity and that DNA shape is a crucial specificity determinant
Professor Rohs can accept graduate students from the following Ph.D. Programs as primary thesis adviser: Computational Biology and Bioinformatics, Molecular Biology, Chemistry, Physics, and Computer Science.

